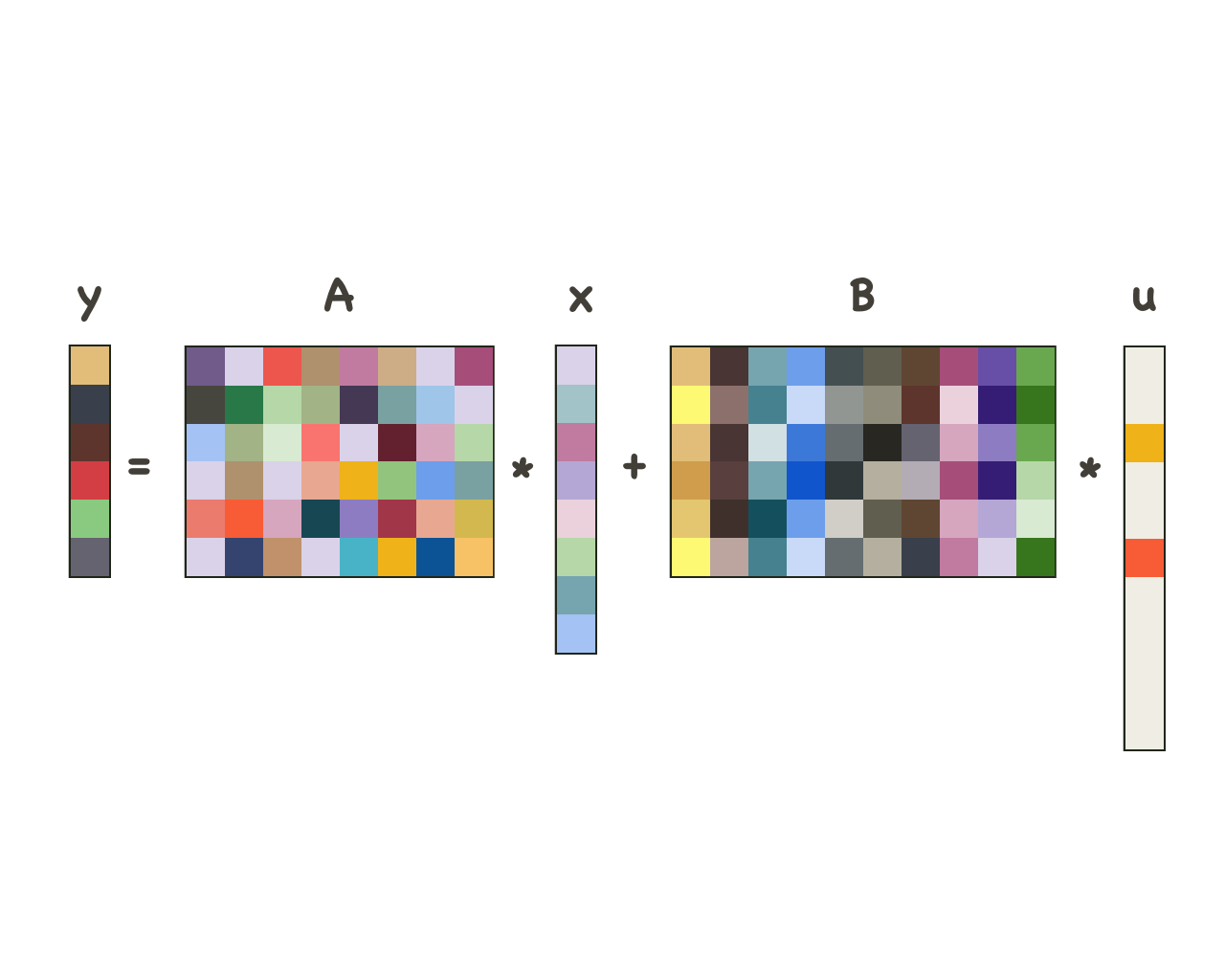~/projects/
Interpretable Artificial and Natural Intelligence
Understanding intelligence—whether biological or artificial—requires reverse-engineering the mechanisms by which systems process information, form representations, and generate novel outputs. Our work in this area develops theoretical frameworks and computational tools to decode the inner workings of neural networks and brains, from uncovering sparse conceptual representations in vision and language models to revealing the geometric principles underlying creative generation in diffusion models. By bridging neuroscience, machine learning, and optimization theory, we aim to build interpretable models that not only explain how intelligence emerges from computation, but also offer new insights into biological cognition and enable the principled design of more transparent AI systems.Current Projects
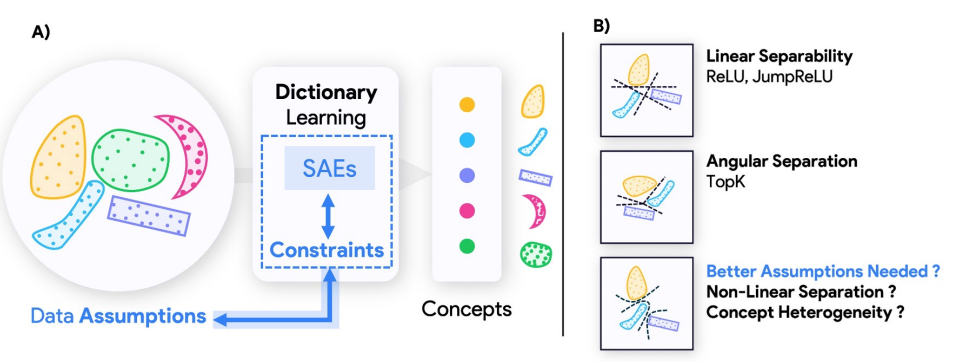
Geometry-Aware Sparse Autoencoders for Mechanistic Interpretability
SAE architectures that extract hierarchical and nonlinearly-separable features for mechanistic interpretability.
SAE architectures that extract hierarchical and nonlinearly-separable features for mechanistic interpretability.
Understanding Vision Transformers Through Dynamics and Geometry
Dynamical systems and geometric tools for decoding how ViTs organize visual information.
Dynamical systems and geometric tools for decoding how ViTs organize visual information.
Understanding Creativity in Diffusion Models
Theoretical frameworks explaining how architectural inductive biases enable creative generation in diffusion models.
Theoretical frameworks explaining how architectural inductive biases enable creative generation in diffusion models.
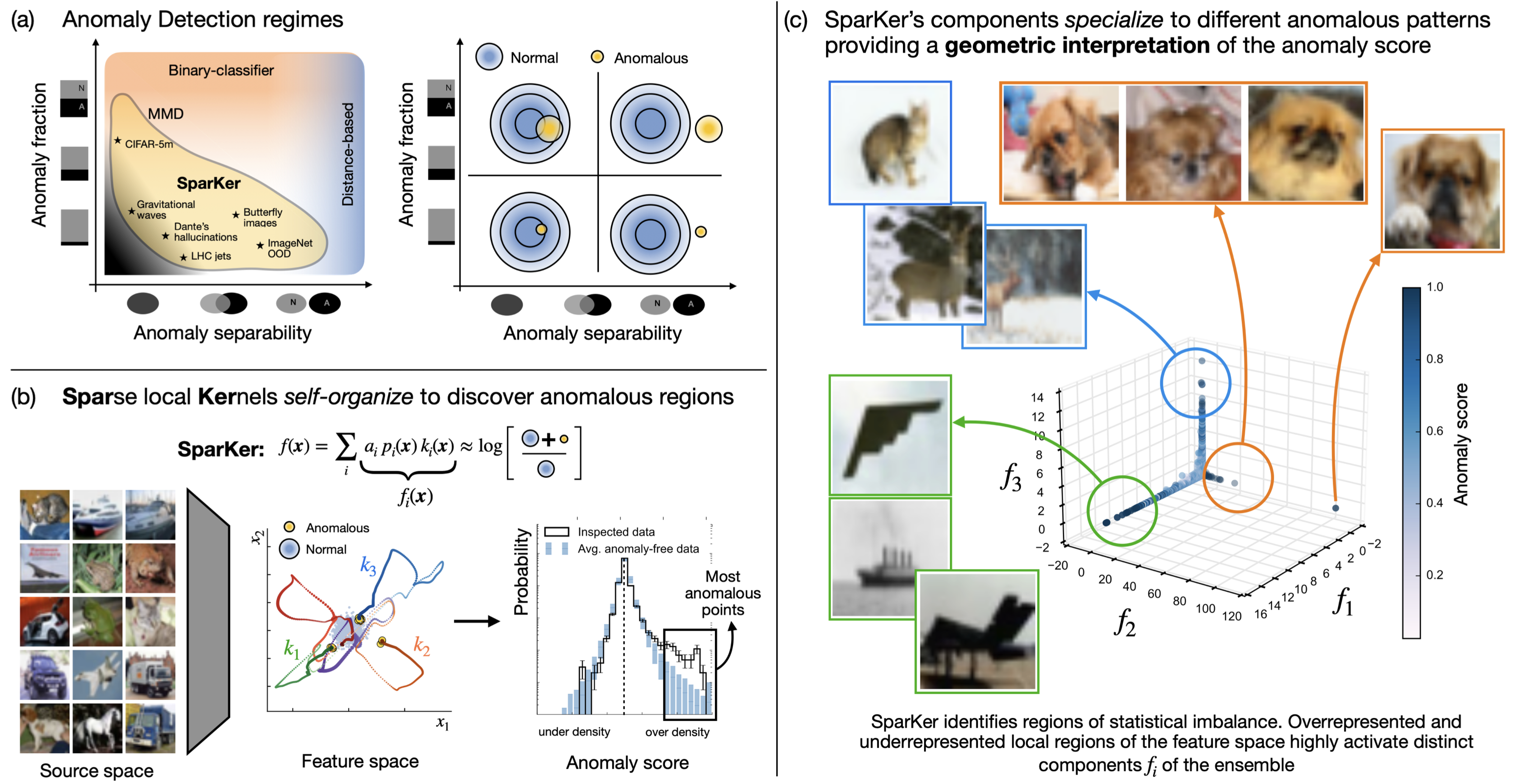
Self-Organizing Anomaly Detection in High-Dimensional Spaces
Sparse, self-organizing Gaussian kernel ensembles that detect rare anomalies in high-dimensional spaces.
Sparse, self-organizing Gaussian kernel ensembles that detect rare anomalies in high-dimensional spaces.
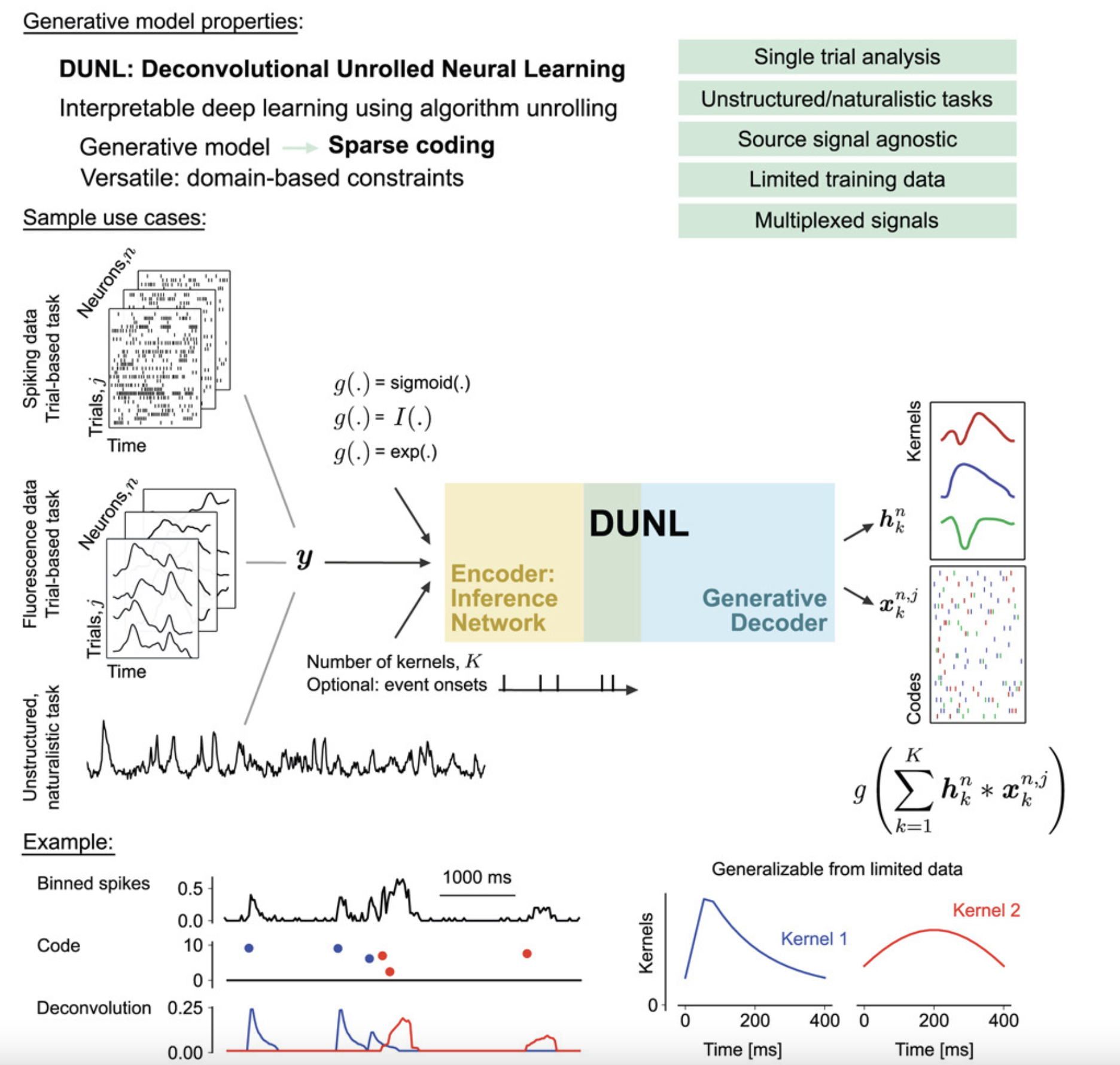
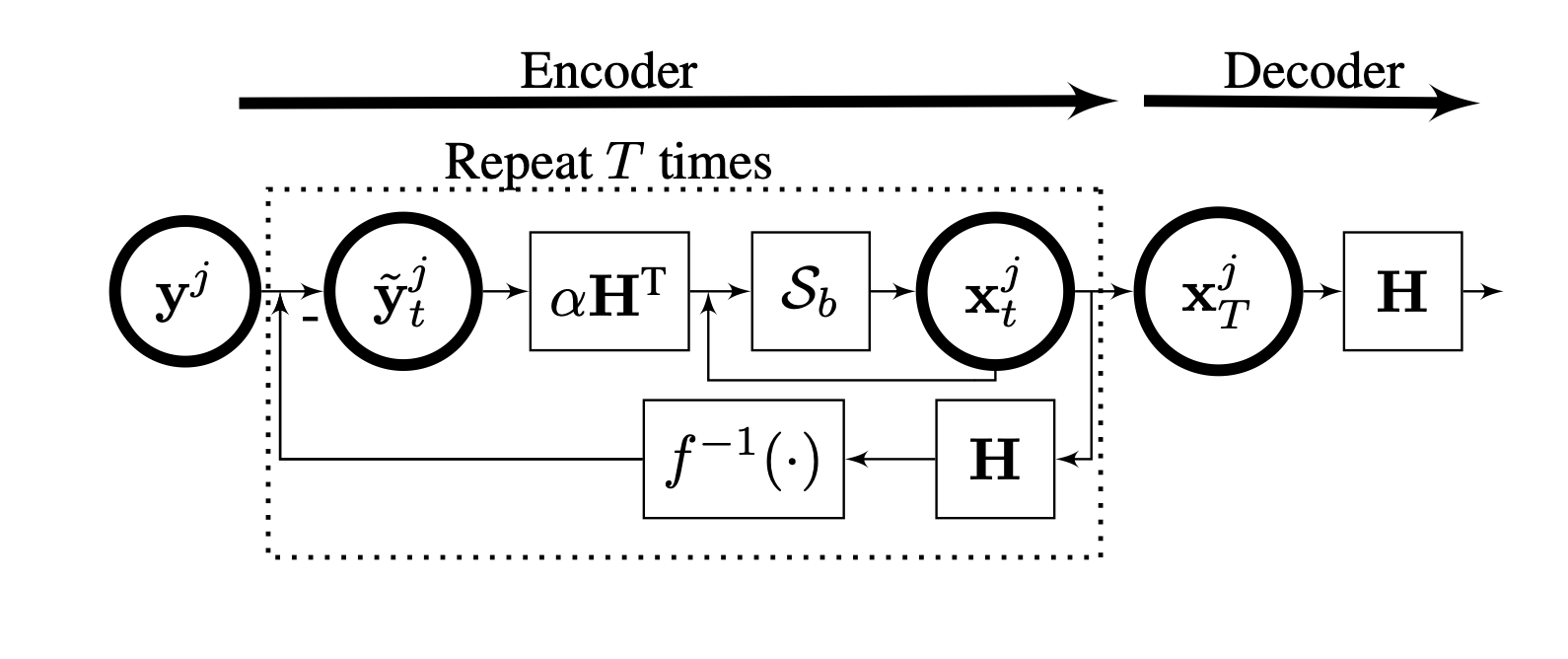
Interpretable Deep Learning for Deconvolutional Analysis of Neural Signals
Algorithm unrolling for interpretable sparse deconvolution of single-trial neural signals across brain areas and recording modalities.
Algorithm unrolling for interpretable sparse deconvolution of single-trial neural signals across brain areas and recording modalities.
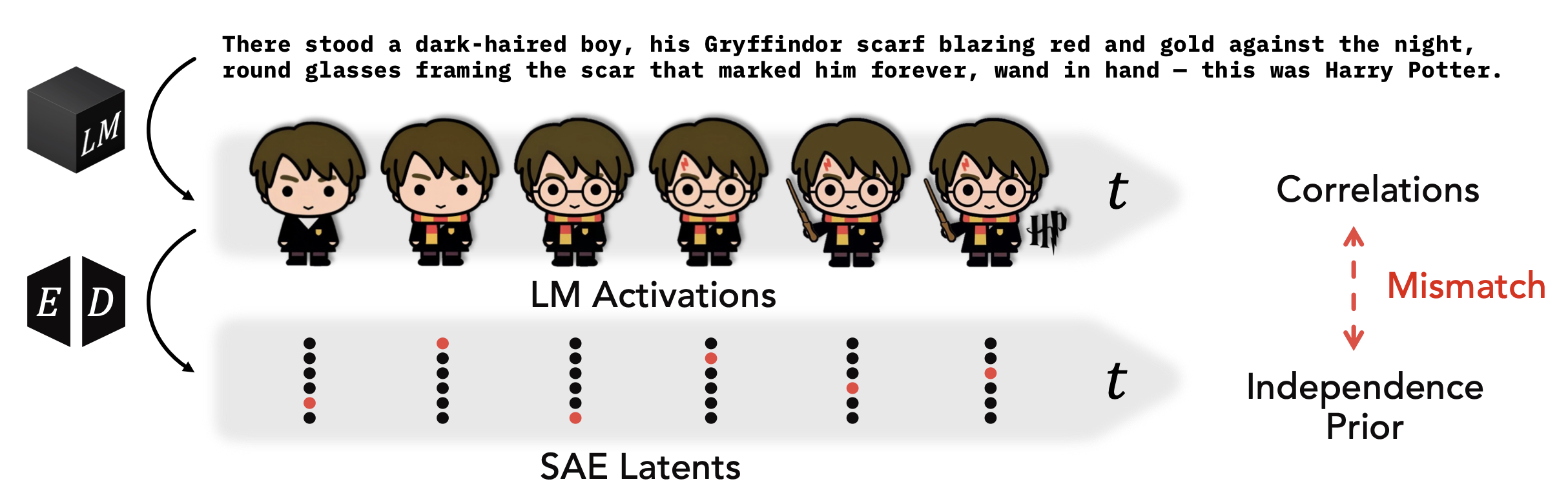
Temporal Structure in Language Model Interpretability
Methods decomposing LM activations into predictable and novel components, capturing temporal dependencies SAEs miss.
Methods decomposing LM activations into predictable and novel components, capturing temporal dependencies SAEs miss.
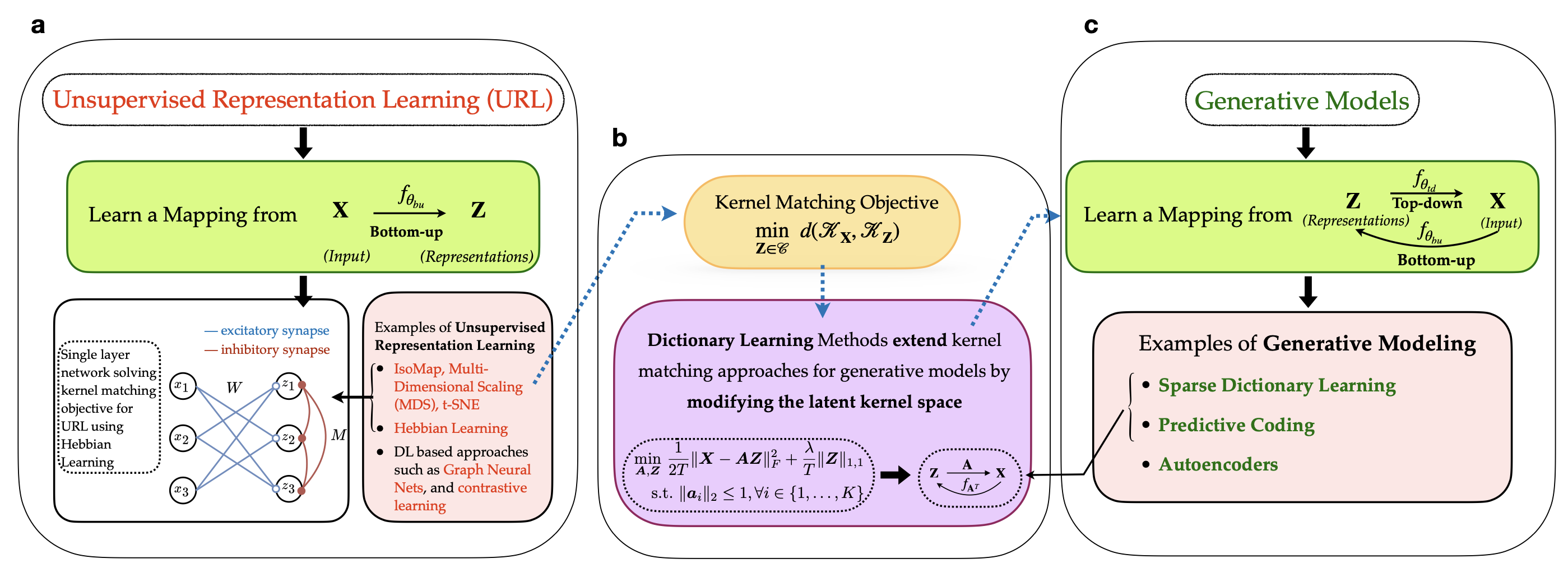
Implicit Generative Modeling by Kernel Similarity Matching
A kernel-based framework for implicit generative modeling that trains without likelihood computation or adversarial objectives.
A kernel-based framework for implicit generative modeling that trains without likelihood computation or adversarial objectives.
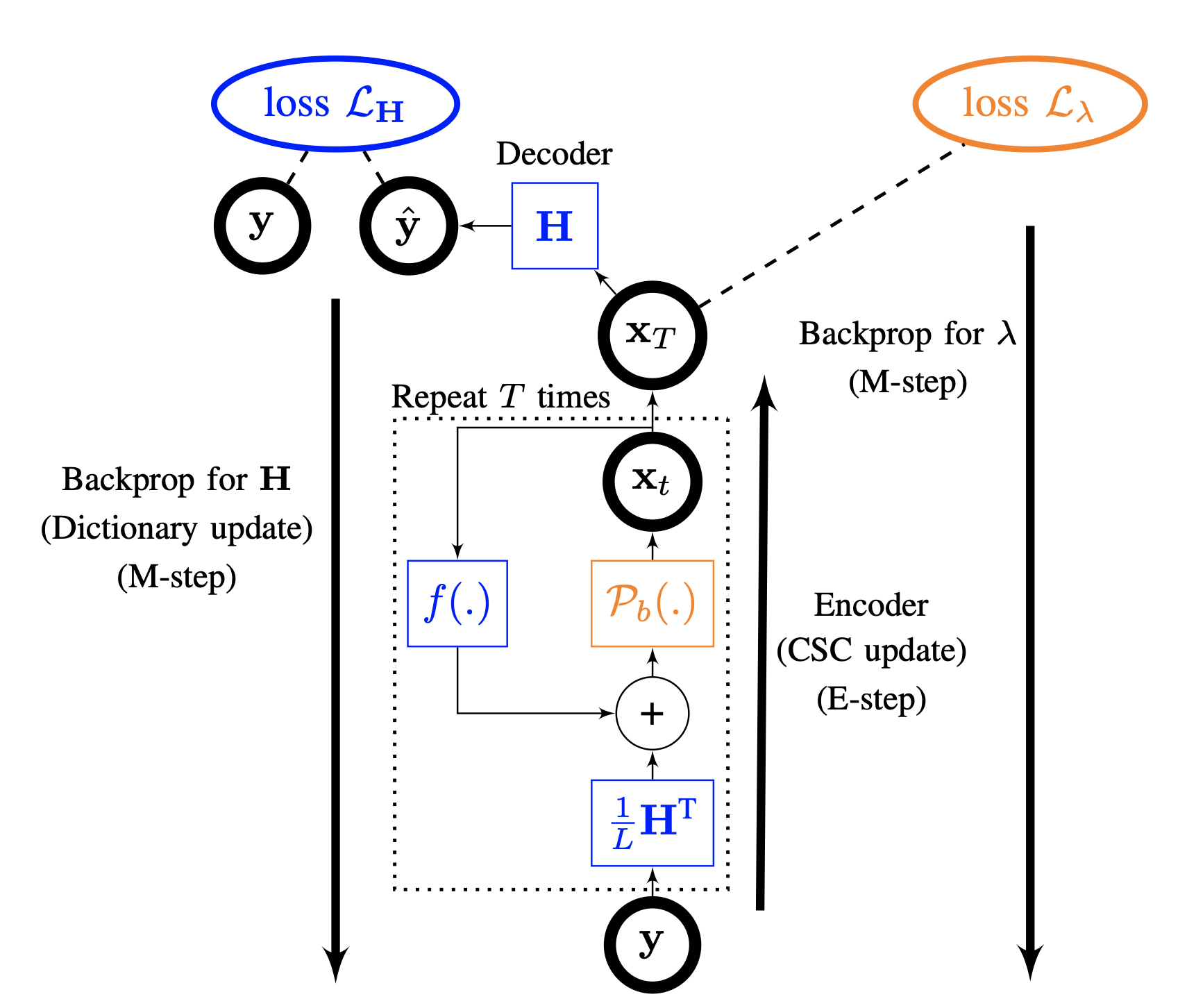
Optimization-inspired Deep Learning
This research uses a process called deep unfolding to show that associated with any mechanistic/generative/statistical model from a large class is a neural network architecture that, when trained, infers the quantities of interest in the model. Stated otherwise, starting from a mechanistic model, deep unfolding lets one design a neural network architecture specifically tailored to the model. On the one hand, the connection to mechanistic models lets us interpret neural networks and enables a theoretical study of their properties, via a study of the properties of the mechanistic model associated with a given architecture. On the other hand, the connection to neural networks lets us leverage GPUs, and the computational infrastructure that has been developed to train neural networks, to solve inference and estimation problems that rely on mechanistic models of data.Current Projects
Implicit Generative Modeling by Kernel Similarity Matching
A kernel-based framework for implicit generative modeling that trains without likelihood computation or adversarial objectives.
A kernel-based framework for implicit generative modeling that trains without likelihood computation or adversarial objectives.
Interpretable Deep Learning for Deconvolutional Analysis of Neural Signals
Algorithm unrolling for interpretable sparse deconvolution of single-trial neural signals across brain areas and recording modalities.
Algorithm unrolling for interpretable sparse deconvolution of single-trial neural signals across brain areas and recording modalities.
Learning Sparse Representations with Matching Pursuit
Replacing SAE encoders with unrolled Matching Pursuit to extract hierarchical sparse representations in highly coherent dictionaries.
Replacing SAE encoders with unrolled Matching Pursuit to extract hierarchical sparse representations in highly coherent dictionaries.
Past Projects
Sparse and dense models
Decomposing signals into low-frequency dense and high-frequency sparse components using a principled computational model with a corresponding deep-learning architecture.
Decomposing signals into low-frequency dense and high-frequency sparse components using a principled computational model with a corresponding deep-learning architecture.
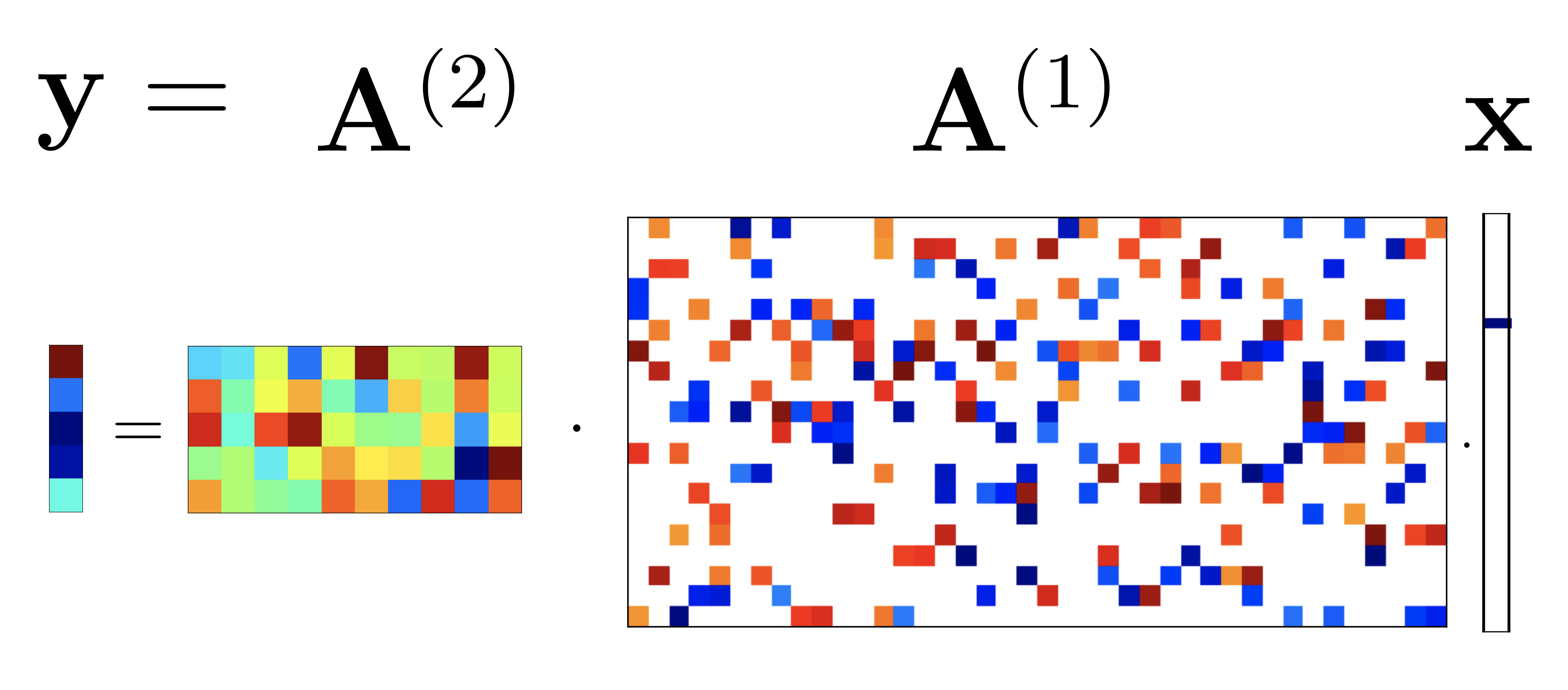
Deeply-sparse signal representations
Theoretical analysis showing deep ReLU networks arise from cascaded sparse-coding models, with provable dictionary recovery and sample complexity bounds.
Theoretical analysis showing deep ReLU networks arise from cascaded sparse-coding models, with provable dictionary recovery and sample complexity bounds.
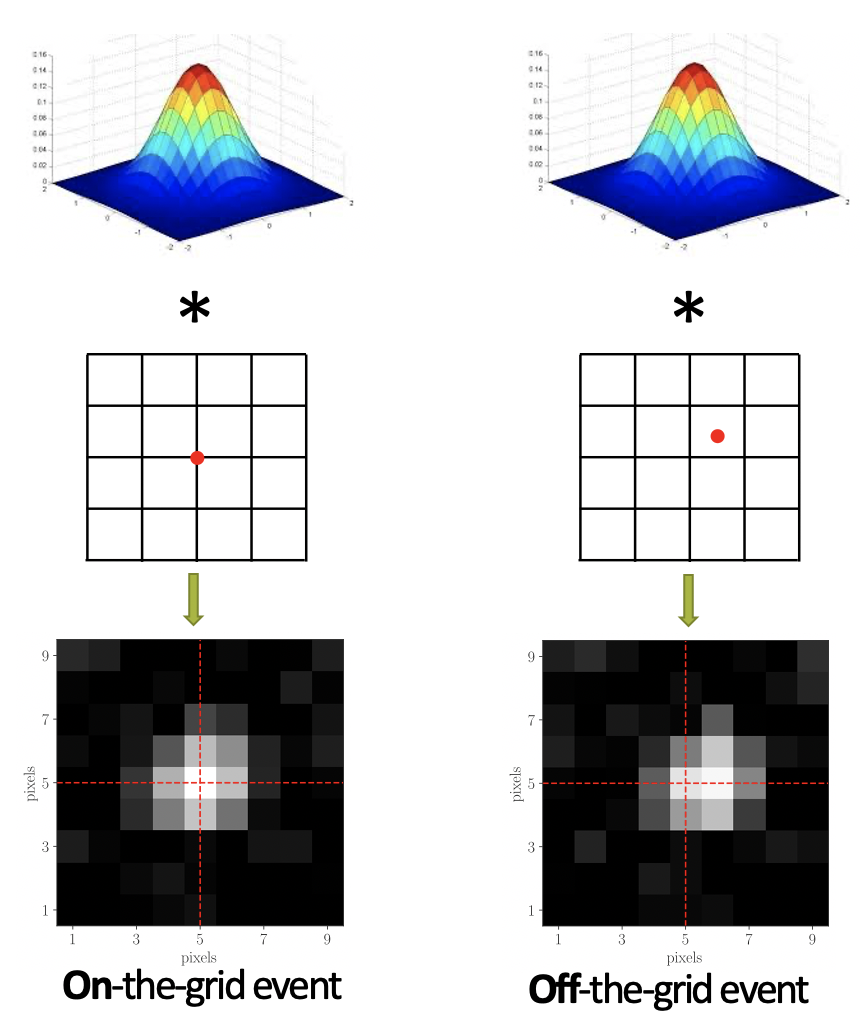
Smoothness-constrained dictionary learning
Incorporating smoothness constraints into dictionary learning to improve element recovery and enable super-resolution in biological signal data.
Incorporating smoothness constraints into dictionary learning to improve element recovery and enable super-resolution in biological signal data.
Learning behaviour of exponential autoencoders
Deep convolutional exponential autoencoders adapted to count, binary, and continuous data, achieving state-of-the-art low-count Poisson denoising.
Deep convolutional exponential autoencoders adapted to count, binary, and continuous data, achieving state-of-the-art low-count Poisson denoising.
Model-based autoencoders for dictionary learning
Constrained recurrent sparse autoencoders (CRsAE) and RandNet for dictionary learning and data recovery from compressed measurements.
Constrained recurrent sparse autoencoders (CRsAE) and RandNet for dictionary learning and data recovery from compressed measurements.
NeuroAI
The brain is a high-dimensional dynamical system that exhibits intricate dynamics at multiple spatial and temporal scales. Our research develops machine learning models inspired by neural computation, bridging neuroscience and artificial intelligence. We investigate how biological mechanisms like traveling waves enable efficient information integration, and develop algorithms for analyzing neural data at heterogeneous spatial and temporal scales. By incorporating biologically-plausible constraints into machine learning architectures, we aim to both understand neural computation and build more efficient AI systems. Our work spans from modeling cortical dynamics to developing data-driven methods for extracting meaningful signals from neural recordings, with applications primarily (but not limited) to neuroscience.Current Projects
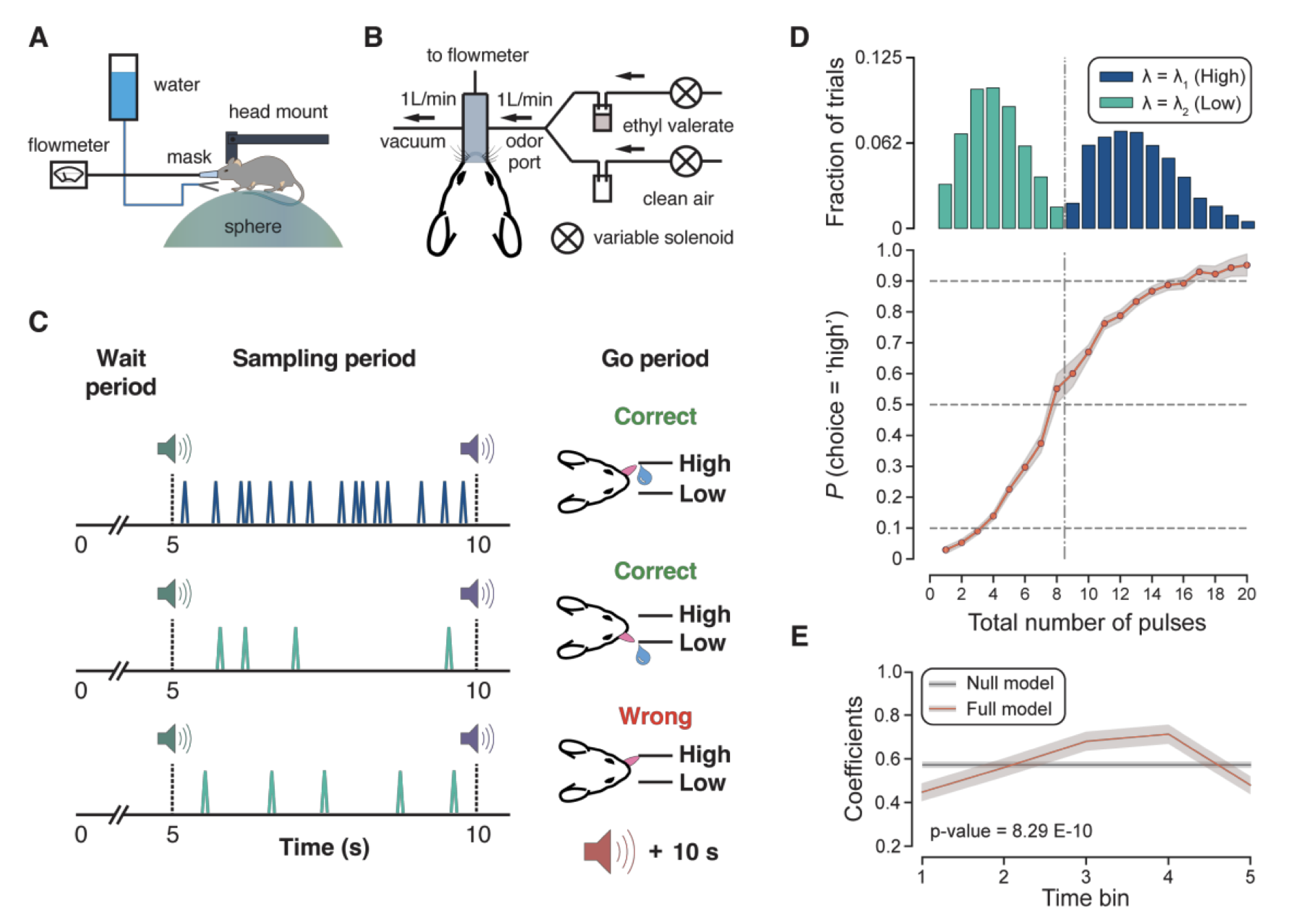
Perception and Neural Representation of Intermittent Odor Stimuli
Neural mechanisms underlying perception of intermittent odor stimuli in mice, characterizing how the brain encodes temporally structured olfactory input.
Neural mechanisms underlying perception of intermittent odor stimuli in mice, characterizing how the brain encodes temporally structured olfactory input.
Traveling Waves for Spatial Information Integration
Convolutional recurrent networks learning traveling wave patterns for long-range spatial information integration in visual tasks.
Convolutional recurrent networks learning traveling wave patterns for long-range spatial information integration in visual tasks.
Implicit Generative Modeling by Kernel Similarity Matching
A kernel-based framework for implicit generative modeling that trains without likelihood computation or adversarial objectives.
A kernel-based framework for implicit generative modeling that trains without likelihood computation or adversarial objectives.
Past Projects
Learning stimuli via dictionary learning
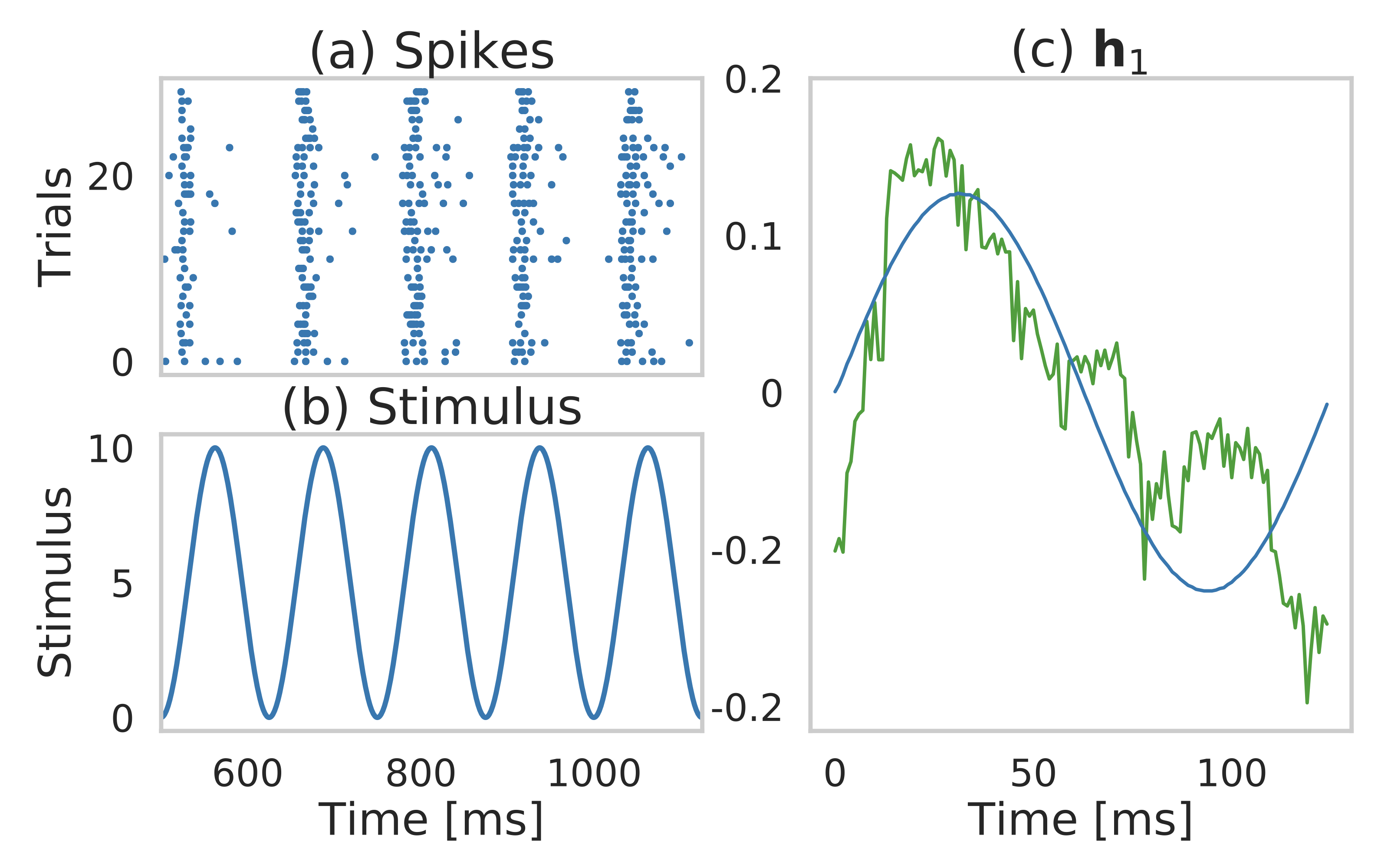
Data-driven dictionary learning from neural spiking data to discover covariates that modulate sensory responses without manual feature selection.
Data-driven dictionary learning from neural spiking data to discover covariates that modulate sensory responses without manual feature selection.
Spike sorting using dictionary learning
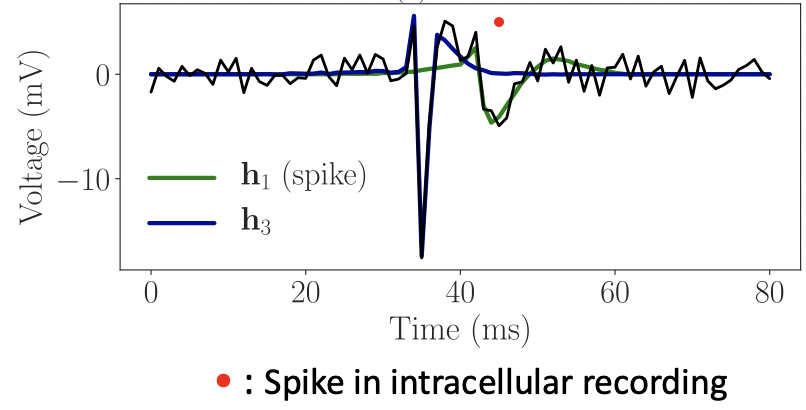
Casting spike sorting as convolutional dictionary learning, yielding more accurate and computationally efficient results than baseline methods.
Casting spike sorting as convolutional dictionary learning, yielding more accurate and computationally efficient results than baseline methods.
Clustering neural spiking data
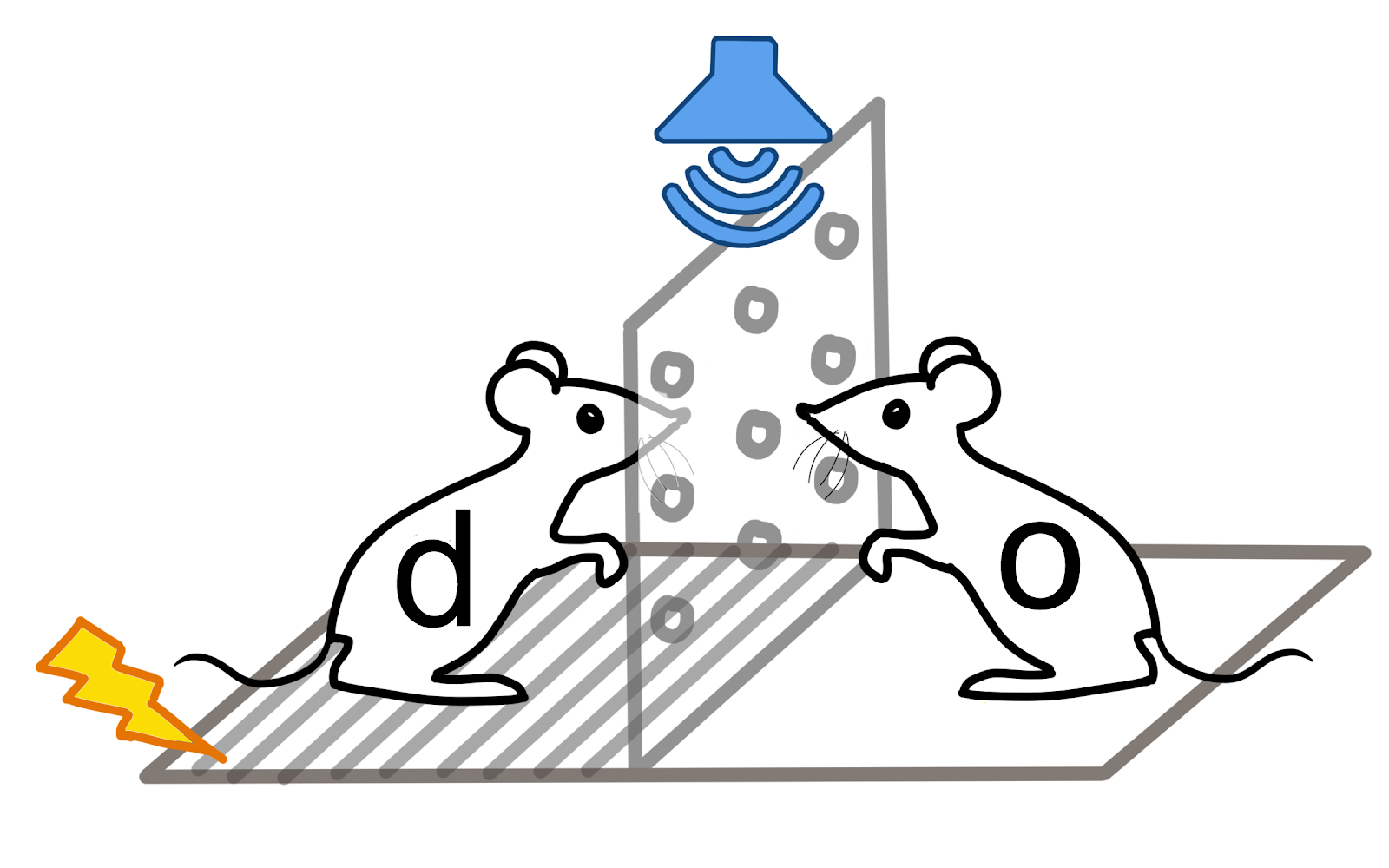
Bayesian nonparametric time-series clustering for discovering grouped neural firing patterns that encode complex behaviors like fear.
Bayesian nonparametric time-series clustering for discovering grouped neural firing patterns that encode complex behaviors like fear.
State-space multitaper time-frequency analysis
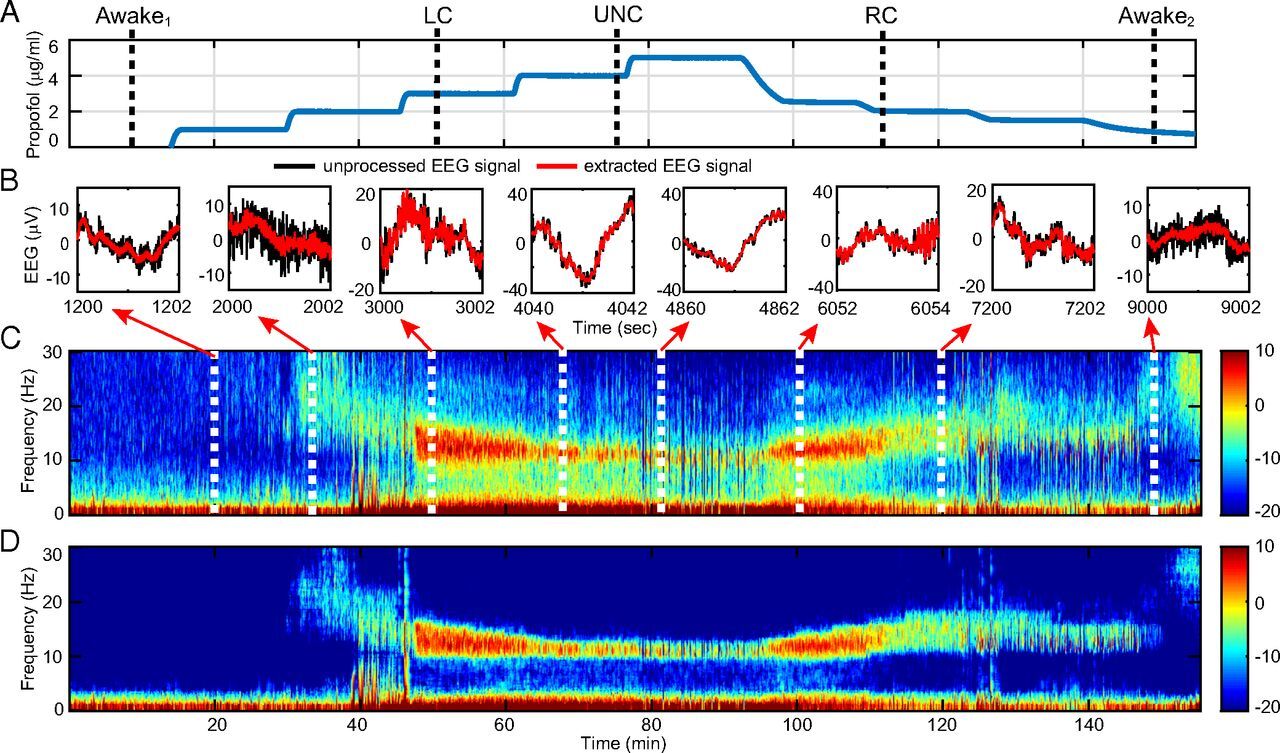
A state-space multitaper framework for nonstationary time-series spectral analysis, providing over 10 dB denoising improvement on EEG recordings.
A state-space multitaper framework for nonstationary time-series spectral analysis, providing over 10 dB denoising improvement on EEG recordings.
Teaching new tricks
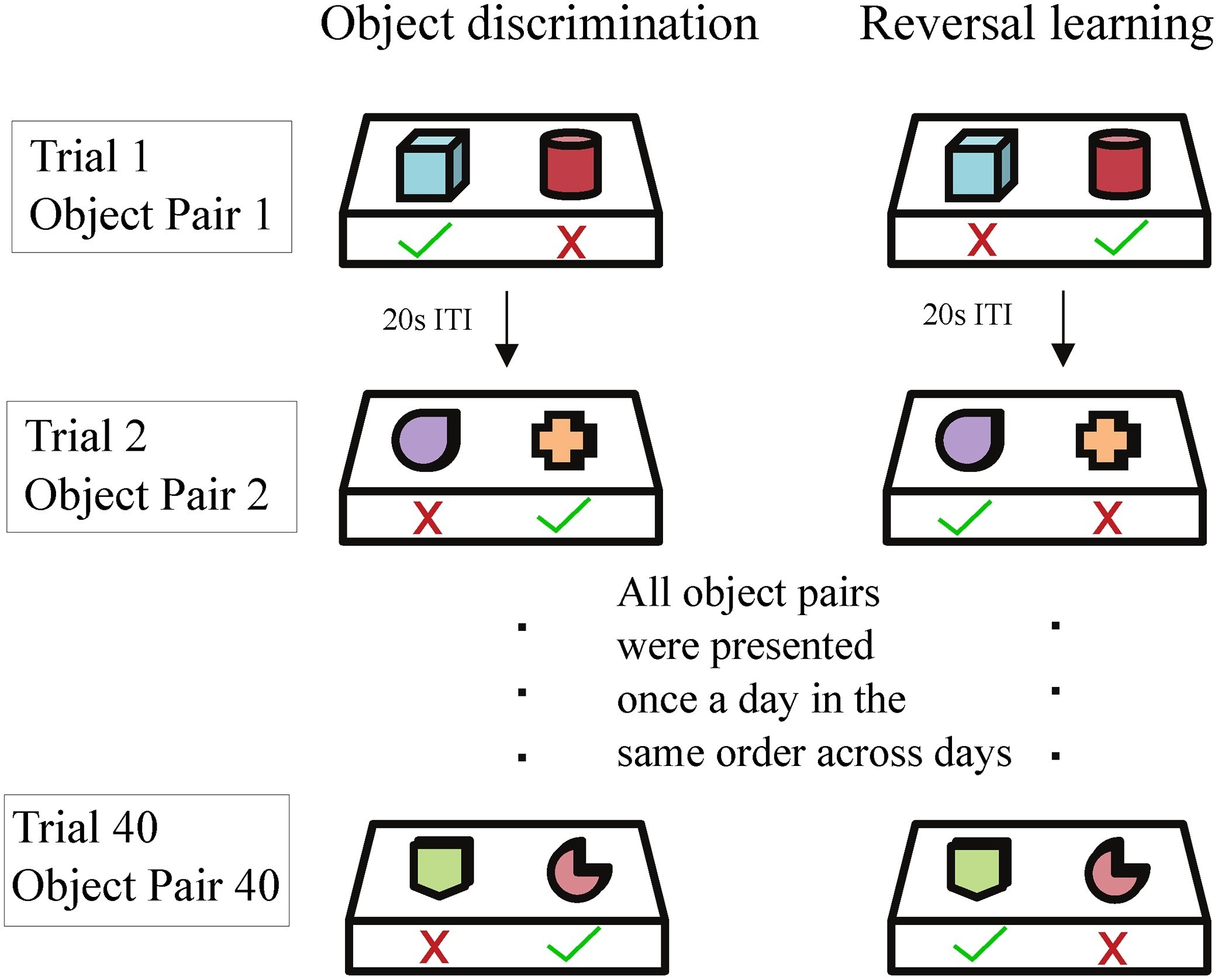
A separable two-dimensional random field model for binary behavioral data that quantifies within-day and across-day learning dynamics in primates.
A separable two-dimensional random field model for binary behavioral data that quantifies within-day and across-day learning dynamics in primates.
Estimating a separably-Markov random field
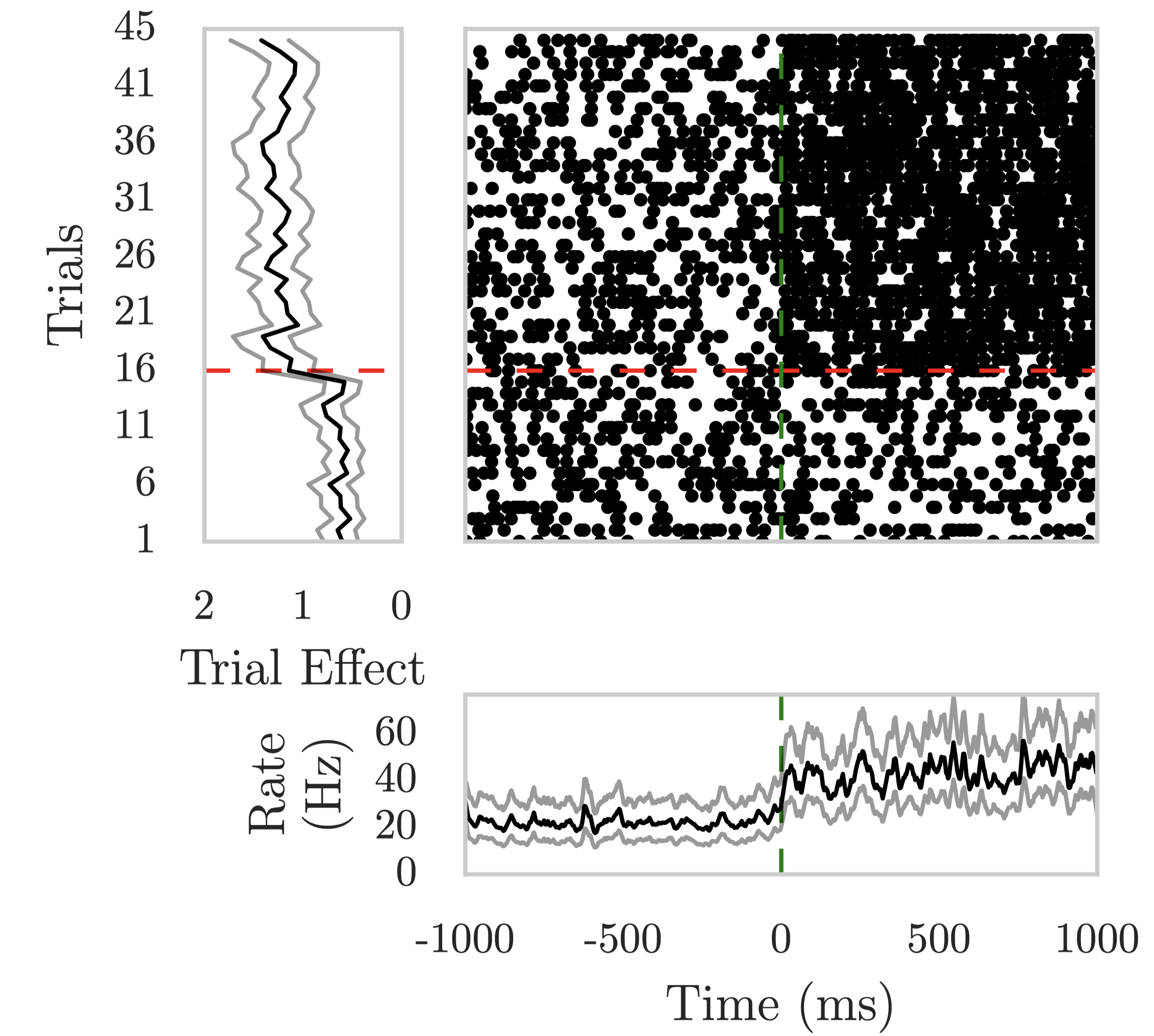
A separable 2D random field model of neural spike rasters that captures trial-to-trial learning dynamics and stimulus-onset latency in fear conditioning.
A separable 2D random field model of neural spike rasters that captures trial-to-trial learning dynamics and stimulus-onset latency in fear conditioning.